DAPD1DatasetsDatabase
Build and maintain the database for the DAP-DataONE Server
Welcome to the DAP-DataONE server database documentation
The DAP-DataONE server is an implementation of a DataONE version 1, tier 1 Member Node. It is designed to act as a kind of broker for DataONE clients and data served using OPeNDAP. The server itself can be found in the project DAPD1Servlet. This project has the code that is used to build, edit and access the database of DAP datasets used by that servlet. The DAP-DataONE sever uses the DatasetsDatabase to hold information each DAP dataset that the server makes available to DataONE clients. The servlet uses information stored in the database to build responses to the various DataONE API calls.
This project is a dependency of the DAPD1Servlet project.
What's here in the documentation?
The documentation contains information about how to:
- Build the software from source
- Testing the software
- Serving your own data
In addition, there is documentation that discusses ways the server could be made more powerful and limitations imposed by the designs of DAP2 and DataONE:
- How it works
- Optimizations and Improvements
Beta software; What we assume
This is beta software without a polished build/install process. We assume you are computer savvy and know how to configure systems, install software, write shell scripts ... all that stuff. If you want to use this software but are unfamiliar with these kinds of things, contact us at support@opendap.org or DataONE:Contact.
Build the software
This project and its companion, DAPD1Servlet, use Maven. Because this project is required by the DAPD1Servlet, if you've followed the build instructions there, you have already built this project. If that's the case, skip down to the section on Testing or on Serving you own data.
Build the code
Use git to clone the project (DAPD1DatasetsDatabase) and build it using mvn clean install. This will build the executable jar file used by the edit-db.sh bash shell script and install that in the local maven repository so it can be found by the DAPD1Servlet build.
Testing the software
The software includes som limited unit tests that will run as part of the maven build, so if the build worked, those passed. This project comes with a test database - one of the advantages of SQLite is that the database files can be moved from machine to machine and still work. To test that the software is working, use the command line tool edit-db.sh to dump the database contents: ./edit-db.sh -d test.db. You should see quite a bit of output:
[nori:DAPD1DatasetsDatabase jimg$] ./edit-db.sh -d test.db
21:46:05.458 [main] DEBUG o.o.d1.DatasetsDatabase.EditDatasets - Starting debug logging
Database Name: test.db
21:46:05.633 [main] DEBUG o.o.d.D.DatasetsDatabase - Opened database successfully (test.db).
Metadata:
Id = test.opendap.org/dataone_sdo_1/opendap/hyrax/data/nc/fnoc1.nc
Date = 2014-07-25T22:26:33.607+00:00
FormatId = netcdf
Size = 24096
Checksum = 1d3afb4218605ca39a16e6fc5ffdfe1544b1dd0a
Algorithm = SHA-1
.
.
.
SMO_Id = test.opendap.org/dataone_smo_1/opendap/hyrax/data/nc/fnoc1.nc
Id = test.opendap.org/dataone_ore_1/opendap/hyrax/data/nc/coads_climatology.nc
SDO_Id = test.opendap.org/dataone_sdo_1/opendap/hyrax/data/nc/coads_climatology.nc
SMO_Id = test.opendap.org/dataone_smo_1/opendap/hyrax/data/nc/coads_climatology.nc
Obsoletes:
[nori:DAPD1DatasetsDatabase jimg$]
Serving your own data
Serving your own data means building a database of datasets using the edit-db.sh tool. Instead of overwriting the test.db database, make a new one. The test.db database is used by the unit tests - those tests depend on its contents. The edit-db.sh tool has online help (-h). Here's a list of its options and how you can use them to build up the database.
The edit-db.sh command always takes the name of a database file. To initialize a new database, use the -i option as in: ./edit-db.sh -i -d my_datasets.db. Note that if you want to erase a database and start over you'll have to delete it using the shell; if you use -i with a database that already exists, you'll get an error.
OK, now with the database initialized, you can use -a to add a dataset like this: ./edit-db.sh -a http://.../fnoc1.nc my_datasets.db. You always need to include the database name; the argument to -a is DAP URL to the dataset. One of the ideas behind DataONE is that datasets can have versions. The database supports this idea by storing a serial number for each version of each dataset. You can edit the database to update the URL for a given dataset using the -u option like this: ./edit-db.sh -u http://.../fnoc1.nc my_datasets.db. This will read new metadata for the URL, bump up the serial number and mark the new information as superseding the old information (DataONE uses the words obsoletes and obsoletedBy). If the URL itself has changes, use the -o option to provide the URL that is being obsoleted by the new URL (the argument to -u). Like this: ./edit-db.sh -u http://.../fnoc2.nc -o http://.../fnoc1.nc my_datasets.db.
At any time ./edit-db.sh -d my_datasets.db will show you the contents of the database.
You can guess that entering each URL by hand could get fairly tiring... but there's an easier way. Use the -r option to read a list of URLs from a file or from standard input. Put the DAP URLs in a file, one per line and use ./edit-db.sh -r datasets.txt my_datasets.db (note that there's a file called dataset.txt in this project and the test.db database can be (re)built using it). The input to -r makes some assumptions about what you want to do. If the URL is already in the databse, it will assume you want to update it (-u); if it's not in the database, it will assume you want to add (-a) it; and if two URLs are given on a line it assumes the first URL should update the second - as if you used -u <first URL> -o <second URL>. Also note that -r will read from standard input if - is used as the file name.
About the design, potential optimizations and its current limitations
This tool, the functions that access the database and the database itself are pretty basic. Information about each dataset is held in six tables in the database. Most of the code that accesses the database is part of this project, shielding the DAPD1Servlet from the actual database structure.
How it works
I'm going to assume you know how DataONE works, at least at a basic level.
For each DAP dataset, there is a single 'base URL' that is used to access both the Science Data Object (SDO) and Science Metadata Object (SMO). The URLs that will return those are built by appending a particular suffix to the base URL. Thus, for each DAP dataset, there is a 'base URL' and two derived URLs. Furthermore, DataONE uses Persistent Identifiers (PIDs) to refer to the SDO, SMO and ORE documents. The PIDs are sent to the DataONE server as an argument of the 'object' function and the DataONE server returns the correct response. Because of this, we must map those PIDs to the URLs that can be used to access the SDO and SMO from the DAP server. This mapping is provided by the database. The ORE document, as explained in the DAPD1Servlet documentation, is stored in the database; more on that in a bit.
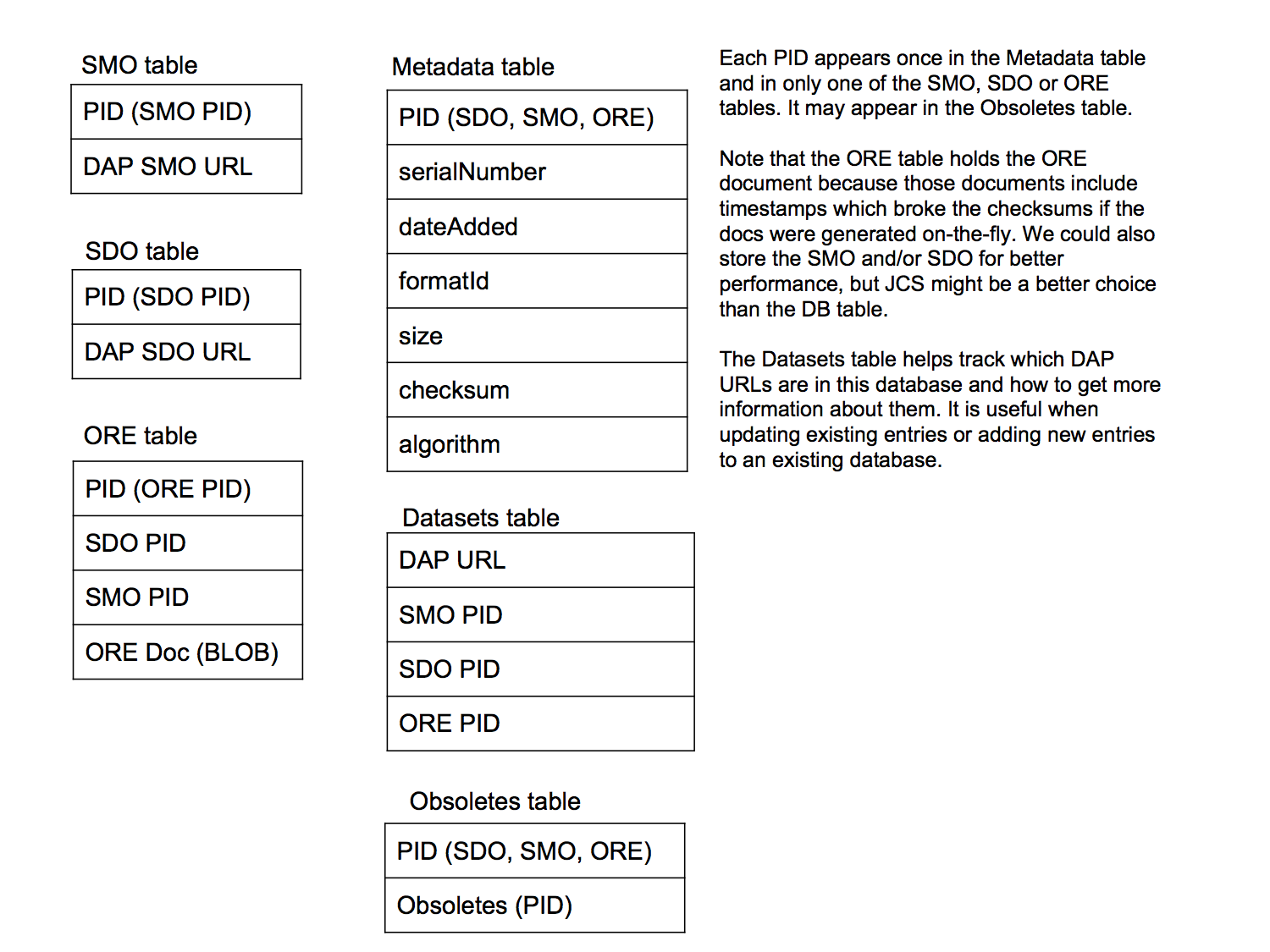
Refer to the figure above. The database uses the SMO, SDO and ORE tables to store information about those kinds of objects - the PIDs that are used to access them and the URL, or the object/response itself for the ORE document, are stored in each of three tables. The Metadata table (creative name ;-) stores information about each PID. The Obsoletes table stores information about datasets that have been updated. Lastly, the Datasets table is used to track DAP URLs and the PIDs that are associated with it. This table is not used by the DAPD1Servlet but is used by the edit-db.sh command line tool to track when a URL has already been added to the database.
Building PIDs
Since DAP datasets are bound to URLs and URLs are unique, it makes sense to use those as the basis of the PIDs used by the server. However, one URL will have several PIDs associated with it - one PID for each of the the SDO, SMO and ORE responses. Then, if the dataset is updated, even if the URL remains the same, there will be a new set of PIDs. The 'formula' for making a PID is to take the DAP URL and build the PID using the host name followed by a token made up of 'opendap', then one of 'sdo', 'smo' or 'ore' and an integer that corresponds to the dataset's DataONE serial number followed by the path part of the DAP URL.
Here's an example PID for a SMO: test.opendap.org/dataone_smo_1/opendap/hyrax/data/nc/fnoc1.nc.
The matching DAP URL is: http://test.opendap.org/opendap/hyrax/data/nc/fnoc1.nc.iso.
Optimizations and Improvements
Because the project also contains code that will be used by the servlet to read from the database, all of the accesses use the JDBC PreparedStatement object. PreparedStatements are precompiled by the underlying JDBC driver and then parameters, if there are any, are substituted as a separate post-compilation step. This prevents SQL injection attacks - an issue with any web interface that uses a database. One optimization would involve reorganizing these PreparedStatements and the objects that used them to minimize the number of times the database is asked to compile them and maximizing the number of times that same objects can be reused.
The database tables are, I think, at least in third normal form and so it is possible to use the SQL JOIN command efficiently to replace multiple queries with a single access to two or more JOINed tables.
It would be much nicer to have a GUI of some sort (a web interface?) that would provide a more interactive way to look at the database contents and update the datasets. At the same time, if the SMO responses were stored in the database this proposed UI could be used to edit/augment the information in those responses. This could improve the performance of the DataONE catalog search functions implemented by the Coordinating Nodes.
Limitations
There is no provision to delete datasets from the database. This is a limitation that comes from the mismatch between DataONE, which provides storage for datasets and can ensure they are never deleted, and DAP which does not. So, the datasets in the database can 'go away' but there's do way to indicate that in the database. Once it's in there ...